PHARMACOGNOSTIC PROFILES AND DNA BARCODING OF THE STEMBARK OF IRVINGIA GABONENSIS (AUBRY-LECOMTE EX O’RORKE) BAIL. (IRVINGIACEAE): A FORENSIC PHARMACOGNOSY DATA
HTML Full TextPHARMACOGNOSTIC PROFILES AND DNA BARCODING OF THE STEMBARK OF IRVINGIA GABONENSIS (AUBRY-LECOMTE EX O’RORKE) BAIL. (IRVINGIACEAE): A FORENSIC PHARMACOGNOSY DATA
Troy Salvia Malgwi, Cletus Anes Ukwubile * and Musa Yusuf Dibal
Department of Pharmacognosy, Faculty of Pharmacy, University of Maiduguri, Nigeria.
ABSTRACT: Irvingia gabonensis is a plant popularly used for both medicinal and dietary purposes. The planthave been used to treat various diseases like diabetes, inflammation, alopecia, fever and ulcers. The study was aimed at evaluating some pharmacognostic fingerprints and molecular DNA barcoding of the stembark. The pharmacognostic fingerprint analysis of the stembark was carried out using standard procedures while the molecular DNA barcoding was done using automated DNA extraction and barcoding by the polymerase chain reaction machine. The organoleptic evaluation showed that the greyish brown stembark of I. gabonensis has a strong bitter taste, acrid odour and slightly coarse to the touch. Chemomicroscopic evaluations reveal the presence of cellulose, suberin, lignin, mucilage, prismatic calcium oxalate crystals and tannins. The transverse section of the stembark showed bicollateral vascular bundle, beaded hexagonal shaped cork cells, fibre sclereids with wide cell lumen, non-striated eccentric oval shaped aggregate starch grains, and wide pith as observed under the scanning electron microscope. The moisture content was 5.13 ± 0.11 w/w %, total ash was 18.58 ± 0.23 w/w/ %, water soluble and acid insoluble ash were13.96 ± 0.66w/v % and 8.02 ± 0.17 w/v % respectively. All the values were within the acceptable ranges. Molecular studies using ribulose biphosphate carboxylase Large chain (rbcL) primers produced consensus DNA that shared alignment and phylogeny with I. gabonensis in the Genbank. Our study showed that I. gabonensis contain important pharmacognostic fingerprints that can distintiguish it from closely related species, and help to check adulteration of drug from the stembark.
Keywords: Pharmacognostic fingerprints, Irvingia gabonensis, DNA barcoding, Polymerase chain reaction, Adulteration
INTRODUCTION: Traditional medicine system continues to play an essential role in health care, with about 80% of the world population relying mainly on traditional medicine as the primary source of health care 1.
It has been well documented that higher plants and their extracts have been used in the treatment of several diseases in traditional African medicine as an age-old practice these plants have been useful in the development of new drugs and continue to play an important role in the drug discovery process therefore there is need to standardize these products 2-3. There is a renewed interest in drugs of natural origin due to their easy availability, accessibility and little or no side effects 4, coupled with a growing interest in correlating the phytochemical constituents of a medicinal plant with its pharmacological activity 5. The Family Irvingiaceae comprises ten species of tropical trees found in Africa, Southeast Asia to western Malaysia. These species usually grow in regions with altitudes from 200 to 500 m with annual rainfall from 1200 to 1500 mm and at temperatures ranging from 20 to 38 °C 6.
They are glabrous trees; wood is extremely hard; leaves sometimes papillate underneath mucilage cells in leaf and stem epidermis having secretory canals containing mucilage in leaves and stems. The leaves are alternate, simple, entire, petiolate, pinnately veined, coriaceous; stipules very large, unequal, intrapetiolar, encircling the terminal bud, early caducous and leaving a very distinct scar. Inflorescences are paniculate, axillary or terminal. The flowers are small, hermaphroditic, regular, pedicels articulated; the sepals are five in number, small and imbricate.
The petals are also five in number, free, imbricate, exceeding the sepals, the stamens are ten in number, which are distinct exceeding the petals and inserted below the large intrastaminal nectary disk. The filaments are plicately folded in bud, the anthers are subbasifixed. The ovary is superior, locular with one ovule per locule. The style is terminal and short. The stigma is punctiform. The fruit is a drupe with one or more pyrenes or a broadly winged samara. The seeds have large embryos, while the cotyledons are flattened and cordate, and the endosperm is fatty. The genera include Desbordesia, Klainedoxa and Irvingia 7.
Irvingia is a genus of African and Southeast Asian trees of the Irvingiaceae family, sometimes known as wild African mango or bush mango. They produce edible mango-like fruits and are prized for their fat and protein-rich nuts. The species are vastly distributed with Irvingia smithii (central Africa), Irvingia gabonensis (west and central Africa), Irvingiagrandifolia (central Africa), Irvingia excels (central Africa), Irvingia malayana (Southeast Asia), Irvingia robur (west and central Africa), Irvingiatenuinucleata (west and central Africa) and Irvingia tenuinucleata (west and central Africa) being the seven (7) species 8-9. Irvingia gabonensis is widely cultivated in West African countries including southwest and southeast Nigeria, southern Cameroon, Côte d'Ivoire, Ghana, Togo, and Benin, to produce its edible fruit whose seed is used in the preparation of local delicious viscous soup for swallowing yam and cassava puddings Oladunjoye and Awani-Aguma 10. Fat extracted from its seeds is commonly known as dika fat and majorly consists of C12 and C14 fatty acids, alongside smaller quantities of C10, C16, and C18, glycerides and proteins.
Irvingia gabonensis seeds are also a good source of nutrients including a variety of vitamins and minerals such as sodium, calcium, magnesium, phosphorus, and iron. It is also a rich source of flavonoids (quercetin and kaempferol), ellagic acid, mono-, di-, and tri-O-methyl-ellagic acids, and their glycosides which are potent antioxidants 11.
Phytochemical analysis of its seeds showed that it contains tannins, alkaloids, flavonoids, cardiac glycosides, steroids, carbohydrates, volatile oils, and terpenoids and its proximate moisture 1.4 ± 0.11%, ash 6.8 ± 0.12%, crude lipid 7.9 ± 0.01%, crude fibre 21.6 ± 0.45%, and crude protein 5.6 ± 0.20% (Mahunu et al., 2019; Olorundare et al., 2020). Pure compounds already isolated from the seed extract include methyl 2-[2-formyl-5-hydroxymethyl)-1 H-pyrrol1yl]-propanoate, kaempferol-3-0-β-D-6″ (p-coumaroyl) glucopyranoside and lupeol (3β-lup-20(29)-en-3-ol). Meanwhile, the antioxidant property of Irvingia gabonensis seed extract has been largely attributed to its high lupeol content 12.
Plant molecular barcoding is a technique used in identifying and classifying plant species based on their DNA sequences. It involves the use of short, standardized DNA sequences, usually from specific genes, as "barcodes" to uniquely identify plant species. The most commonly used DNA sequences for plant molecular barcoding are those from the chloroplast genome, specifically the regions known as ribulose bisphosphate carboxylate large gene (rbcL) and Maturase K gene (mat K). These regions are highly conserved, meaning they are similar across plant species, but also contain enough variation to differentiate between different plant species 13. The process of plant molecular barcoding typically involves several steps. First, plant tissue is collected and DNA is extracted from the sample. The rbcL and mat K regions of the chloroplast genome are then amplified using polymerase chain reaction (PCR), a process that selectively amplifies specific DNA sequences. The PCR products are then sequenced, and the resulting DNA sequences are compared to a reference database of known plant barcodes to identify the plant species 14.
Plant molecular barcoding has several advantages over traditional methods of plant identification, such as morphology-based identification. DNA sequences are less subject to variation due to environmental factors, making molecular barcoding more reliable and accurate. Additionally, DNA sequences can be easily stored and shared in databases, facilitating the creation of a comprehensive library of plant barcodes for future use 15.
Irvingia gabonensis is known to contain a variety of bioactive compounds with potential health benefits. However, there are concerns about the sustainability of harvesting the bark of this tree for medicinal use, as well as the potential for misidentification and adulteration of different samples of bark. By using DNA barcoding techniques, researchers can accurately identify different samples of Irvingia gabonensis stembark and distinguish them from other plant species. This can help to ensure that the bark is being harvested sustainably and that consumers are getting the genuine product. It can also help to prevent the sale of adulterated or counterfeit products 16.
Several methods can be used for DNA barcoding of Irvingia gabonensis stembark, including PCR amplification of specific regions of the DNA and sequencing of the amplified DNA. Once the DNA sequences have been obtained, they can be compared to a reference database of known sequences to identify the species.
DNA barcoding of Irvingia gabonensis stembark can provide a valuable tool for ensuring the authenticity and sustainability of this important medicinal resource. It has the potential to improve the quality and safety of traditional medicines and commercial products, as well as aid in forensic investigations 17. However, to the best of our knowledge, there is no report on the pharmacognostic fingerprints and DNA barcoding of the stembark of I. gabonensis. Therefore, the present study was aimed at identifying some pharmacognostic profiles of stembark and providing its DNA barcoding for easy taxonomic identification from closely related species which will help in assessing adulteration of medicinal and economic products from the stembark of the plant.
MATERIALS AND METHODS:
Chemicals, Reagents, and Equipment: Molecular and analytical tools such as cetyltri-methylammonium bromide buffer (CTAB), Proteinase- K, R-Nase A, 95% ethanol, isopropanol, DNA polymerase (Taq polymerase), deoxyribonucleoside triphosphates (dNTPs), PCR primers (rbcL II), DNA polymerase (e.g., Taq polymerase), sequencing primers (specific to the barcoding region of interest), fluorescent dyes, PCR machine (Eppendorf master-cycler nexus X2 thermal cycler), UV-vis spectrophotometer r(NANODROP 2000c Thermo-Scientific, UK), gel electrophoresis system (Maxi-Broad-H), centrifuge (Eppendorf 5702), refrigerator and freezer (Haier Thermocool Refrigerator HR-130 AS –Silver/Haier Thermocool Chest Freezer HTF 150HAS R6 Silver), Vortex mixer (LABOID L 20), UV trans-illuminator Visualizer (Enduro-temp V1) and DNA extraction kits Table 1.
TABLE 1: DNA EXTRACTION KIT SPECIFICATIONS USED
| Component | Product procured (mL) | Volume used (mL) |
| Lysis buffer L | 1 x 30, 2 x 60 | 30 |
| Binding buffer I | 1 x 7, 1 x 25 | 7 |
| Solution WN | 1 x 18, 1 x 55 | 18 |
| Wash solution A | 2 x 38 | 38 |
| Elution buffer B | 1 x 30 | 15 |
| RNase A | 5 vials (5 x 80 µL) | 1 vial (80 µL) |
| Spin columns | 1 x 250 | 50 |
| Filter columns | 1 x 250 | 50 |
| Collection tubes | 1 x500 | 100 |
| Elution tubes | 1 x 250 | 50 |
| Product insert | 1 x 1 | 1 |
Collection, Identification, and Preparation of Plant Materials: Fresh stembarks of I. gabonensis were collected from a forest in the Iragbiji community, Osun State, Nigeria, in October 2018. The plant was identified by a taxonomist Mr Namadi Sunusi of the Department of Botany, Ahmadu Bello University, Zaria.
A voucher specimen number ABU06926 was deposited for the plant at the herbarium of the Department of Botany for referencing. The stembarks were allowed to dry under shade for two weeks and prepared accordingly following standard procedures.
Macroscopic examination of I. gabonensis Stembark: Macroscopic observations were carried out on the whole dried stembark of the plant, and these entail colour, odour, taste, texture, shape and fracture 18.
Microscopic analysis of I. gabonensis Stembark: To remove obscuring materials, the powdered stembark of I. gabonensis was cleared with a 70% chloralhydrate solution and boiled in a water bath for 30 min. The cleared sample was mounted on a microscopic slide with dilute glycerol and observed under the microscope (40x), for the presence of pharmacognostic fingerprints of the powdered stembarks. Suitable photographs were taken and recorded 19.
Scanning Electron Microscopy of the Leaf of I. gabonensis: The detailed internal structure (transverse section) of the leaf was observed using a Phenom desktop scanning electron microscope (PhenomWorld, USA) at 8000x magnification to reveal some pharmacognostic features.
Chemo-microscopic Analysis on I. gabonensis Stembark: This was carried out on powdered stembark of I. gabonensis. Briefly, the finely crushed stembark was cleared using 70% chloral hydrate solutionto remove obscuring materials. It was then heated for 30 minin a water bath. Using two drops of dilute glycerol, the cleared sample was then placed on a microscope slide, and the presence of several cell inclusions and cell wall components was observed using various detecting reagents 19-20.
Determination of Physicochemical Parameters of I. gabonensis Powdered Stembark: The physicochemical parameters such as moisture content, ash value, total ash, extractive values, acid insoluble, water-soluble ash values, swelling and foaming indices where determined following standard procedures. For each parameter, a total of ten replicate readings were taken, and results were recorded as mean ± SE.
DNA Barcoding of I. gabonensis Stembark: DNA barcoding of I. gabonensis was carried out according to the methods by Kress et al., 15 and de Oliveiraet al., 21. Various steps were involved following protocols previously described.DNA extraction was carried out using the DNA kit (NORGEN BIOTEK© Plant DNA isolation kit, USA).
DNA Extraction Procedure:
Lysate Preparation: Less than 100 mg of plant tissue was placed into a mortar and ground into a powder. The ground material was then transferred into a DNase-free 1.7 mL- micro-centrifuge tube with 500 µL of lysis buffer L, and 1µL of RNAse A was added. This was incubated at 650 C for 10 min with occasional mixing of the lysate 3 times during incubation by inverting the tube. 100µL of binding buffer 1 was then added, mixed thoroughly and incubated for 5 min on ice.
The filter column (clear O-ring) with one of the provided collection tubes was then assembled, and the lysate was pipetted into the filter column and spun for 2 min at 20,000 x g (~14,000 RPM). The clear supernatant from the flow-through was then transferred into a DNAase-free micro-centrifuge tube using a pipette. An equal volume of 70% ethanol was added to the lysate collected (100µL of ethanol was added to every 100µL of lysate), and vortex to mixed 21.
Binding to Column: A spin column (grey O-ring) with a collection tube was assembled, and then up to 650µL of the clarified lysate with ethanol onto the spin column and centrifuge for 1 min at 10,000 x g (~10,000 RPM). The flow-through was discarded, and then the spin column with the collection tube was reassembled.
Column Wash: 500µL of solution WN was added to the column and centrifuge for 1 min at 20,000 x g (~14,000 RPM). The flow-through was discarded and the spin column with its collection tube was reassembled. 500µL of Wash solution A was added to the column and centrifuge for 1 min at 20,000 x g (~14,000 RPM).
The flow-through was then discarded and the spin column with its collection tube was reassembled, and the process repeated. The column was spun for 2 min at 20,000 x g (~14,000 RPM) to thoroughly dry the resin, and the collection tube was discarded.
DNA Elution: The column was placed into a fresh 1.7 mL elution tube, and 100 µL of elution buffer B was added to the column and incubated for 1 min at room temperature, then centrifuged for 1-2 min at 10,000 x g (~10,000 RPM).
DNA Quantification: The extracted DNA was then analysed using a UV-vis spectrophotometer (NANODROP 2000c), and the concentration of DNA was then obtained.
DNA Storage: The purified genomic DNA extracted was then stored in a DNA sample bottle at 2-8 0 C in a refrigerator.
DNA Amplification: The polymerase chain reaction (PCR) master mix containing the I. gabonensis DNA, the forward primer, reverse primer, deoxynucleotide triphosphates (dNTPs) mix, taq polymerase, buffer solution with DNA polymerase and magnesium chloride (MgCl2) solution was placed in a PCR tube and vortex gently to ensure homogeneity.
The rbcL II primer was used in this mix. The PCR tubes were then placed into the thermal cycler, ensuring they were securely sealed; then the appropriate heating block was set on the PCR machine. After the thermal cycling process was completed, the tubes were removed and run on agarose gel with a DNA ladder to estimate the size of the amplified product 21.
Agarose Gel Electrophoresis: This procedure was used to separate and analyze the DNA obtained from thermocycler 15.
Briefly, 2 g of agarose powder was weighed and added to a beaker containing 200 mL volume of TAE/TBE buffer, to make (1% w/v) the mix was then gently heated on a hot plate until the agarose was completely dissolved. 0.5 μg/mL ethidium bromide was the added to the warm agarose solution.
The solution was mixed well and poured into the gel casting tray, a space was left for the comb and was allowed to cool and solidify. The DNA sample and DNA ladder were then mixed with bromophenol blue which is the loading dye and then kept ready. The comb was carefully removed from the solidified gel and placed into the gel electrophoresis apparatus filled with TAE/TBE buffer, ensuring the wells were on the cathode side. Using a micropipette, the DNA sample and ladder were carefully placed into separate wells.
The electrophoresis was run at 90V for 45 min. After this, the gel was removed and placed on a UV transilluminator where the separated DNA fragments were visualized and recorded 21.
DNA Sequencing and Analysis: The purified DNA fragment obtained was then sequenced; where the sequence obtained was trimmed and cleaned to remove any low-quality or ambiguous bases. The consensus sequences were then subjected to sequence analysis using bioinformatics tools; comparing with reference sequences available in public DNA barcode databases GenBank and BOLD (Barcode of Life Data Systems).
Statistical Analysis: Data obtained from the physicochemical analysis were expressed as mean± SE (n = 10).
RESULTS:
Macroscopic Parameters of I. gabonensis Powdered Stembark: The organoleptic evaluation of I. gabonensis stembark showed a brown to greyish colour powder with a slightly acrid odour and strong bitter taste as well as coarse texture Table 2.
TABLE 2: ORGANOLEPTIC PARAMETERS OF I. GABONENSIS POWDERED STEMBARK
| Test | Observation |
| Colour | Greyish-brown |
| Odour | Acrid |
| Taste | Strong bitter |
| Texture | Coarse |
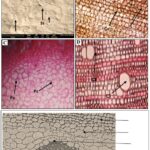
Microscopic Examination of I. gabonensis Stembark: The scanning electron microscopy of the leaf revealed the presence of a few numbers of paracytic stomata on the adaxial surface (upper) than on the abaxial surface (lower) which is typical of most terrestrial plants among the angiosperms Fig. 1.
FIG. 1: SCANNING ELECTRON MICROSCOPY OFA LONGITUDINAL SECTION THROUGH I. GABONENSIS SHOWING PARACYTIC STOMATA (ARROWS); 8000X (A), BEADED CORK CELLS (B), PARENCHYMATOUS CELLS; PC STAINED WITH SAFRANIN (C), FIBRE SCLEREIDS WITH WIDE CELL LUMEN; CL (D), AND TRANSVERSE SECTION OF STEMBARK WITH WIDE PITH (E) AS VIEWED USING THE SCANNING BIOLOGICAL MICROSCOPE, 3000X. AFEW STOMATA WERE OBSERVED ON THE UPPER SURFACE OF THE LEAF TYPICAL OF TERRESTRIAL ANGIOSPERMS (A).
Chemomicroscopical Evaluation of the Powder Stembark of I. gabonensis: Chemomicroscopical evaluation of the powdered stembark of I. gabonensis revealed the presence of cellulose cell wall, lignified cell wall, tannins, starch, calcium oxalate, suberin and mucilage Table 3.
TABLE 3: CHEMOMICROSCOPICAL FEATURES OF I. GABONENSIS STEMBARK POWDER
| Constituents | Inference |
| Starch | + |
| Lignin | + |
| Tannins | + |
| Calcium oxalate | + |
| Calcium carbonate | - |
| Cellulose | + |
| Suberin | + |
| Gums and Mucilage | + |
+: present and -: absent.
Evaluation of Physicochemical Parameters of I. gabonensis the Stembark: The total ash was 18.58 ± 0.23% w/w, acid-insoluble ash was 8.02 ± 0.17% w/w and water- soluble ash was 13.96 ± 0.66% w/w, to the physicochemical parameters. The ethanol-soluble extractives were 18.20 ± 0.28% w/v and 28.60 ± 0.12 % w/v water-soluble extractives. The moisture content was 5.13 ± 0.11. The plant's swelling index was 4.55 ± 0.32 mL. The plant's foaming index was calculated as 142.9± 0.61% w/v Table 4.
TABLE 4: PHYSICOCHEMICAL PARAMETERS OF I. GABONENSIS STEMBARK POWDER
| Parameters | Values (%w/w or w/v) ± SE |
| Moisture content | 5.13 ± 0.11 |
| Total ash | 18.58 ± 0.23 |
| Acid insoluble ash | 8.02 ± 0.17 |
| Water soluble ash | 13.96 ± 0.66 |
| Ethanol extractive | 18.20 ± 0.28 |
| Water extractive | 28.60 ± 0.12 |
| Swelling index | 4.55 ± 0.32 mL |
| Foaming index | 142.9± 0.61 |
Data were expressed as means ± SE (n = 10).
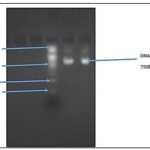
DNA Molecular Barcoding Analysis: Results of the gel electrophoresis after visualization show a clear band concerning the ladder as presented in Fig. 2.
FIG. 2: ELECTROPHORESIS GEL STAINED WITH BROMOPHENOL BLUE UNDER UV TRANSILLUMINATOR COMPARED WITH A 1200BP LADDER
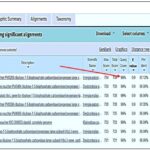
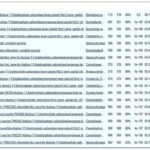
Sequencing and Analysis of DNA Fragments: Fig. 3(A) below shows the consensus DNA after the sequencing process. This sequence was obtained after the purified fragment was trimmed and cleaned to remove any low-quality or ambiguous bases. The consensus DNA obtained was analysed with bioinformatics tools; the DNA was compared with reference sequences available in public DNA barcode databases NCBI and blasted. Fig. 3(B) and (C) show that the consensus sequence has an alignment like that of I. gabonensis. Similarly, Fig. 3(D) shows the consensus DNA’s phylogenic relationships.
AGCAAGTGTTGGATTCAAAGCTGGTGTTAAAGATTATAAATTACTTATTATACTCCTGACTATGAAACCAAAGATACTGATATCTTGGCAGATTCCGAGTAACTCCTCAACCTGGAGTTCCGCCTGAGGAAGCAGGGGCGCGTTGCTTCTACTGGGACATGGACAACTGTGTGGACCGGGACTTACCTAGTCTTGATCGTTATAAGGGAAGATGCTACCATCCCGTTGCGGTGGAGAAGAAAATCAATATATTGCTTATGTAGCTTATCCTTTCCTGAACTAAATGTTTACTTCCATTGTGGGTAATGTATCGGGTTGGCAAAGTCTACGCGCCCTGCGTTTGGAGGATTTGCGAATCCCTGCCTACTGCTTATACTAAAACTTTCCAAGGTCCGCCTCATGGCATTCAAGATGAACAAGTACGGTCGCCCCCTATTGGGCTGACTATAACCTAAATTGGGTTTATCCGCTAAGAATTACGGTAAGTTTATGAATGTCTCCGCGGTGGGCTTGATTTTACGAAAGATGATGAGATGTGGGTACGTGAATTCCCAACCATTTATGCGTTGGAGAGACCGTATTTTGTGCCGAAGCGCTTTTTAAAGCACAGGCCGAAACAGGTGAAAGACA TTATTTGAAT GCTACTCGTC
FIG. 3: CONSENSUS DNA OBTAINED FROM SEQUENCING OF THE DNA FRAGMENTS BY THE PCR MACHINE(A), CONSENSUS DNA MATCHING WITH I. GABONENSIS SPECIES IN THE GENBANK (B), CONSENSUS DNA ALIGNMENT WITH SPECIES OF I. GABONENSIS PRESENT IN THE GENBANK GRAPHICS (C), AND PHYLOGENY TREE SHOWING THE TAXONOMIC RELATIONSHIP BETWEEN IRVINGIA SPECIES AND THE CONSENSUS DNA FRAGMENTS(D). THE RED ARROW INDICATES THE TAXON OF I. GABONENSIS FROM THE GENBANK.
DISCUSSION: Macroscopical evaluation of the stembark of I. gabonensis reveals a greyish-brown colouration. The stembark is known for its distinctive greyish-brown colouration. This colouration is a result of tannins and flavonoids that are present in the plant, which was observed and reported by Ojo 22. Tannins are a type of plant compound that is often responsible for the astringency or bitterness of certain foods 23. The organoleptic evaluation of the stembark of I. gabonensis showed that the bark is greyish-brown in colour. The colour of the stembark is usually associated with the age of the plant, as it matures plant naturally takes up a brownish coloration 24-25. Environmental conditions are also implicated in the colour of the stembark as described by Savage and Vellend 26. Other factors such as the composition of minerals in the soil have also proven to be significant as a determinant of the colour of the stembark 27.
This research has also discovered that the odour of the stembark of I. gabonensis is acrid, this was also described in a research conducted in 2020 by Egwunatum, and co-workers 28, to study the effect of variegated forest soil amendments on the germination and early growth of I. gabonensis where the odour of the stembark was one of the parameters evaluated and was concluded to be acrid. Otitolaiyeet al. 29, in research conducted to evaluate the phytochemical activity and in-vitro antioxidant potential of aqueous and ethanol extracts of I. gabonensis stembark, reported the odour as bitter. The odour of I. gabonensis stem bark can be associated with the presence of resins in the cells 30.
A study conducted by Godayol et al. 31, which aims to study odour-causing organic compounds in wastewater treatment plants, reported that the presence of certain phenols and aldehydes in the stem bark which they postulated is responsible for its odour. Kola-Mustapha et al. 32, reported that environmental conditions play a significant contribution to the odour that is associated with stembarks. The texture of I. gabonensis as determined by this research is coarse, this was also the conclusion drawn by Nuhu and co-workers in 2020 33, in research that aimed to establish the macroscopic profile of I. gabonensis in Wistar rats. Cell division that is influenced by the environment can affect the texture of the stembark Funada et al. 34. A tough fibrous sclerenchyma can also be implicated in the texture of the stembark, this was described in research conducted by Zhang, and co-workers 35. The chemomicroscopic analysis of the stem bark of I. gabonensis revealed the presence of cellulose, suberin and lignin, this is consistent with research conducted by Nuhu et al. 33 while investigating the safety profile of I. gabonensis rootbark extract. Mgbemena, and co-workers in 2019 36, studying the chemical composition, proximate and phytochemical analysis of the peels, seed coat, leaves and seeds of I. gabonensis, reported the presence of cellulose, suberin and lignin in their chemomicroscopic analysis.
Cellulose typically adds to the structural support of the plant hence its presence is integral for strength and rigidity Pauly et al. 37, this discovery was made while studying the molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Cellulose also serves as a barrier against pathogens that could damage tissues involved in the transportation of water and minerals Souza et al. 38. Therefore cellulose plays a significant role in regulating water and nutrient exchange in the plant, this is made possible through small pores present in them that assist in the movement Franca etal. 39. Suberin which was present in the stembark is a waxy hydrophobic substance that helps to prevent water loss in the plant especially when environmental conditions are not favourable de Silva et al. 21. Suberin also protects from pathogens and extends the lifespan of the plant Shukla and Barberon 40. Like cellulose, suberin found in the stem bark plays an important role in regulating the movement of water and nutrients in and out of plant tissues. Baxter et al. 41.
A moisture content of 5.13%for the stembark of I. gabonensis indicates that 5.13% of the weight of the bark is water. Moisture content is an important parameter in the processing and storage of plant materials. A high moisture content can lead to microbial growth and spoilage, while a low moisture content can result in brittleness and loss of quality. The ideal moisture content for the stembark of I. gabonensis would depend on the intended use of the material. For medicinal use, a moisture content of 5.13% is relatively low and indicates that the bark has been properly dried and stored. This low moisture content helps to preserve the active compounds in the bark and ensures its effectiveness. All these and other pharmacognostic fingerprints evaluated in this study perform various functions in plants and are used in the pharmaceutical industry for quality assurance.
The obtained DNA sequence of 650 base pairs after sequencing was subjected to a BLAST (Basic Local Alignment Search Tool) analysis. BLAST is a widely used bioinformatics tool that compares the query sequence against a vast database of known sequences to find similarities. The BLAST analysis has shown that the 650-base pair sequence from I. gabonensis aligns significantly with an entry in the GenBank database. This suggests that the sequence obtained from the organism shares similarities with a previously documented sequence in GenBank. The fact that the sequenced DNA aligns with a known sequence in the GenBank database supports the accuracy and validity of the sequencing process. It indicates that the DNA sample extracted from I. gabonensis is consistent with genetic information previously documented for this species. While the alignment with an existing sequence is a positive indication, further analysis may be required to fully characterize the genetic makeup of I. gabonensis. This will involve sequencing comparison with additional sequences, and functional analysis of specific genes or regions.
CONCLUSIONS: Our study comprehensively evaluated various pharmacognostic parameters and DNA barcoding of I. gabonensis stembark, shedding light on its botanical and genetic characteristics to provide accurate taxonomic identification. Through detailed analysis, essential parameters such as moisture content, acid-insoluble ash, and total ash among others were determined, providing crucial insights into the plant's quality and purity. Moreover, the successful extraction and sequencing of a DNA fragment from the chloroplast gene further enriched our understanding of the genetic makeup of I. gabonensis. The deposition of this DNA sequence in GenBank enhances accessibility for future studies and facilitates accurate species identification and authentication. By integrating traditional pharmacognostic methods with modern molecular techniques, this research contributes valuable data to the field of Pharmacognosy and genetic resource conservation. Moving forward, the findings from this study serve as a foundation for further research into the medicinal and genetic potential of I. gabonensis.
ACKNOWLEDGEMENTS: We are thankful to the Biotechnology Center University of Maiduguri, Nigeria for their numerous helps in DNA analysis.
Funding: None.
Ethical Consideration: Not applicable to this aspect of research.
CONFLICT OF INTEREST: We have none.
REFERENCES:
- Baxter I, Hosmani P, Rus A, Lahner B, Borevitz J, Muthukumar B and Salt D: Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. Journal of Genetics 2009; 5(5): e1000492.
- Brain KR and Turner TD: The practical evaluation of phytopharmaceuticals. Wright - science-technica, Bristol 1975; 36-82.
- Chanda S: Importance of pharmacognostic study of medicinal plants: An overview. Journal of Pharmacognosy and Phytochemistry 2014; 2(5): 69-73.
- Cowan R and Fay M: Challenges in the DNA barcoding of plant material. Plant DNA Fingerprinting and barcoding: methods and protocols 2012; 23-33.
- Cragg GM, Newman DJ and Snader KM: Natural products in drug discovery anddevelopment. J Nat Prod 1997; 60(1): 52-60.
- de Silva ND, Murmu J, Chabot D, Hubbard K, Ryser P, Molina I and Rowland O: Root suberin plays important roles in reducing water loss and sodium uptake in Arabidopsis thaliana. Metabolites 2021; 11(11): 735.
- Egwunatum AE, Dolor DE and Ofobike CJ: Effect of variegated forest soil amendments on the germination and early growth of Irvingia gabonensis (O Rorke, Baill). Asian J. of Res in Agriculture and Forestry 2020; 6(1): 43-50.
- Ekpe OO, Nwaehujor CO, Ejiofor CE, Arikpo PW, Woruji EE and Amor ET: Irvingia gabonensis seeds extract fractionation, its antioxidant analyses, and effects on red blood cell membrane stability. PhOL 2019; 1: 337–353.
- Evans WC: “Trease and Evans pharmacognosy”, 16th edition, W. B. Saunders Ltd., London, UK 2011; 191-393.
- Fajar A, Ammar GA, Hamzah M, Manurung R and Abduh MY: Effect of tree age on the yield, productivity, and chemical composition of essential oil from Cinnamomumburmannii. Current Research on Bioscences and Biotechnology 2019; 1(1): 17-22.
- Farnsworth NR and Fabricant DS: The value of plants used in traditional medicine for drug discovery. Environmental health perspectives 2001; 109 Suppl. 1(Suppl 1), 69–75. doi:10.1289/ehp.01109s169.
- Franca D, de Barros JRS and Faez R: Spray-dried cellulose nanofibrils microparticles as a vehicle for enhanced efficiency fertilizers. Cellulose 2021; 28: 1571-1585.
- Funada R, Yamagishi Y, Begum S, Kudo K, Nabeshima E, Nugroho WD and Nakaba S: Xylogenesis in trees: from cambial cell division to cell death. In Secondary xylem biology 2016; 25-43, Academic Press.
- Godayol A, Alonso M, Besalú E, Sanchez JM & Anticó E: Odour-causing organic compounds in wastewater treatment plants: Evaluation of headspace solid-phase microextraction as a concentration technique. J Chromatography A 2011; 1218(30): 4863-4868.
- Harris DJ and Leaky TM: A revision of the Irvingiaceae in Africa. Bulletin du Jardin Botanique National de Belgique 1996; 65(1–2): 143–196.
- Hollingsworth PM, Graham SW & Little DP: Choosing and using a plant DNA barcode. PloS one, 2011; 6(5): e19254.
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA & Janzen DH: Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences 2005; 102(23): 8369-8374.
- Kubitzki K: Irvingiaceae. In Flowering Plants. Eudicots 2014; 229-232. Springer, Berlin, Heidelberg.
- Kola-Mustapha AT, Taiwo SO, Isiaka AR, Amao SO, Ishola IO, Ghazali YO & Usman SO: Evaluation of Terminalia macroptera (Combretaceae) Guill. & Perr stem bark extract incorporated into an emulgel for the potential management of rheumatoid arthritis. Scientific African 2023; 19: e01557.
- Leakey R & Newton A: Domestication of tropical trees for timber and non-timber products. MAB Digest 1994; 17: 67–68.
- Lev-Yadun S: Why is latex usually white and only sometimes yellow, orange or red? Simultaneous visual and chemical plant defense. Chemoecology 2014; 24: 215-218.
- Li DZ, Liu JQ, Chen ZD, Wang H, Ge XJ, Zhou SL & Chen SL: Plant DNA barcoding in China. Journal of Systematics and Evolution 2011; 49(3): 165-283.
- Mgbemena NM, Ilechukwu I, Okwunodolu FU, Chukwurah JVO & Lucky IB: Chemical composition, proximate and phytochemical analysis of and peels, seed coat, leaves and seeds. Ovidius University Anls Chem 2019; 30(1): 65-69.
- Nuhu A, Abdurahman EM, Danmalam UH, Kawu MU, Zakariya AM & Ayeni AE: Safety profile of Irvingia gabonensis (Aubry-Lecomte ex O’Rorke) Baill. root bark extract: Acute and sub-acute toxicity studies in Wistar rats. J Med Plants for Econ Dev 2020; 4(1): 8.
- Ojo OA: Preliminary Qualitative Analysis for Cancer Chemopreventive Agents in Irvingia gabonensis Baill, AlstoniaBoonei De Wild. and BrideliaferrugineaBth. Stem Bark from South West, Nigeria. Afr J of Basic & AppL Sci 2014; 6(4): 110-114.
- Okolo CO, Johnson PB, Abdurahman EM, Abdu-Aguye I & Hussaini IM: Analgesic effect of Irvingia gabonensis stem bark extract. J Ethnopharmacol 1995; 45(2): 125-129.
- Oladunjoye AO and Awani-Aguma EU: Effect of thermosonication on physicochemical, bioactive, sensory and microbiological qualities of African mango fruit (Irvingia gabonensis) juice. Measurement: Food 2023; 100103.
- Otitolaiye C, Omonkhua A, Oriakhi K, Okello E, Onoagbe I & Okonofua F: Phytochemical analysis and in-vitro antioxidant potential of aqueous and ethanol extracts of Irvingia gabonensis stem bark. Pharmacognosy Research 2023.
- Owolabi J, Omogbai EKI and Obasuyi O: Antifungal and antibacterial activities of the ethanolic and aqueous extract of Kigella africana (Bignoniaceae) stem bark. Afr J Biotechnol 2007; 6: 882-885.
- Patil UH & Gaikwad DK: Effect of varying environmental conditions of mineral status of stem bark of Anogeissus latifolia. J Pharm Res 2012; 5(2): 1140-43.
- Pauly M, Albersheim P, Darvill A & York WS: Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. The Plant Journal 1999; 20(6): 629-639.
- Raj SP, Solomon PR & Thangaraj B: Irvingiaceae. In: Biodiesel from Flowering Plants 2022; 365-369. Springer, Singapore.
- Savage JA & Vellend M: Elevational shifts, biotic homogenization and time lags in vegetation change during 40 years of climate warming. Ecography 2015; 38(6): 546-555.
- Sharma A, Flores-Vallejo RDC, Cardoso-Taketa A and Villarreal ML: Antibacterial activities of medicinal plants used in Mexican traditional medicine. J Ethnopharmacol 2017; 208: 264-329. doi: 10.1016/j.jep.2016.04.045. Epub 2017 May 4.
- Shukla V and Barberon M: Building and breaking of a barrier: Suberin plasticity and function in the endodermis. Current Opinion in Plant Biology 2021; 64: 102153.
- Souza A, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C and Somerville SC: Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiology 2017; 173(4): 2383-2398.
- Sun J and Chen P: Ultra-high-performance liquid chromatography with high-resolution mass spectrometry analysis of African mango (Irvingia gabonensis) seeds, extract, and related dietary supplements. J. of Agric. and Food Chem 2012; 60(35): 8703–8709. doi: 10.1021/jf302703u.
- Vijayan K & Tsou CH: DNA barcoding in plants: taxonomy in a new perspective. Current Science 2010; 1530-1541.
- WHO, Quality Control Methods for Medicinal Plants. WHO, Geneva, Switzerland 2011; 9-31.
- Wolfe OA and Ijeoma UF: Effects of aqueous extracts of Irvingia gabonensis seeds on the hormonal parameters of male guinea pigs. Asian Pac. J Tro Med 2010; 3(3): 200-204.
- Zhang W, Xue Y, Yang S, Wang Y & Zhao H: Sclereids are strong enough to support the delicate corollas: experimental and computational data evidence from Camellia sinensis (L.). Scientific Reports 2017; 7(1): 1-7.
How to cite this article:
Malgwi TS, Ukwubile CA and Dibal MY: Pharmacognostic profiles and DNA barcoding of the stembark of Irvingia gabonensis (aubry-lecomte ex o’rorke) bail. (Irvingiaceae): a forensic pharmacognosy data. Int J Pharmacognosy 2024; 11(6): 303-14. doi link: http://dx.doi.org/10.13040/IJPSR.0975-8232.IJP.11(6).303-14.
This Journal licensed under a Creative Commons Attribution-Non-commercial-Share Alike 3.0 Unported License.
Article Information
10
303-314
2366 KB
740
English
IJP
Troy Salvia Malgwi, Cletus Anes Ukwubile * and Musa Yusuf Dibal
Department of Pharmacognosy, Faculty of Pharmacy, University of Maiduguri, Nigeria.
doccletus@yahoo.com
02 June 2024
26 June 2024
29 June 2024
10.13040/IJPSR.0975-8232.IJP.11(6).303-14
30 June 2024